[2025-04] Our lab participated in the 19th Annual Breast Center Retreat, gave oral and poster presentations, and won the group karaoke competition! 
[2025-03] Lihong Xu from the Immunology & Microbiology Program joined the group for a research rotation. Welcome, Lihong!
[2025-03] Faye’s paper Deciphering the dark cancer phosphoproteome using machine-learned co-regulation of phosphosites has been published in Nature Communications. Congratulations! This study introduces CoPheeMap, a network built from phosphoproteomic data across 11 cancer types that maps how over 26,000 phosphorylation sites are co-regulated. Using this network, we further developed CoPheeKSA, a machine learning model that predicts which kinases modify which phosphosites. The model uncovered over 24,000 kinase–substrate relationships, including many involving poorly understood sites and kinases. Validated with experimental data, these tools help reveal cancer-related signaling changes and highlight potential new drug targets. The code for CoPheeMap and CoPheeKSA is available on GitHub at: https://github.com/bzhanglab/CoPheeMap.
[2025-02] Congratulations to Michael for passing the CCB PhD Qualifying Exam!
[2025-01] CCB student Shiyi Wu joined the group for a research rotation. Welcome, Shiyi!
[2025-01] QCB student Fanwei Ruan joined the group for a research rotation. Welcome, Fanwei!
[2024-12] Holiday party at Topgolf. 
[2024-12] The paper Mapping the functional network of human cancer through machine learning and pan-cancer proteogenomics has been published in Nature Cancer. Congratulations to co-first authors Zhiao and Jonathan! By applying supervised machine learning to extensive proteomics and RNA sequencing data from 1,194 individuals spanning 11 cancer types, this study constructs a functional network, FunMap, comprising 10,525 protein-coding genes. FunMap achieves unprecedented precision in linking functionally associated genes, surpassing traditional protein-protein interaction maps. Analysis of FunMap identifies functional protein modules, reveals a hierarchical structure linked to cancer hallmarks and clinical phenotypes, provides deeper insights into established cancer drivers and predicts functions for understudied cancer-associated proteins. Additionally, applying graph-neural-network-based deep learning to FunMap uncovers drivers with low mutation frequency. The FunMap Python package is fully open source and available for download from the Python Package Index (https://pypi.org/project/funmap). The source code is hosted on GitHub (https://github.com/bzhanglab/funmap). In addition, a web application, accessible at https://funmap.linkedomics.org/, offers visualization tools to explore gene neighborhoods, dense modules, and the hierarchical organization of this pan-cancer FunMap.
[2024-12] James successfully defended his PhD thesis, congratulations, Dr. Moon! He will be joining Sage Bionetworks next month. Best wishes for a bright future!
[2024-11] Dr. Moran Chen joined the lab as a postdoctoral research associate. Welcome, Moran!
[2024-11] Xinpei’s paper Tumor-associated antigen prediction using a single-sample gene expression state inference algorithm has been published in Cell Reports Methods. This study introduces a Bayesian-based algorithm for inferring gene expression states in individual samples. By integrating this algorithm into a computational workflow for tumor-associated antigen (TAA) identification, the study uncovers TAAs shared across tumors within and between cancer types, including a repository of experimentally validated peptides to support further immunotherapy development for liver cancer.
[2024-10] Congratulations to Jonathan on starting his new role as an Instructor at the Breast Center!
[2024-08] The paper Pan-cancer proteogenomics expands the landscape of therapeutic targets has been published in Cell! Congratulations to co-first authors Sara, Xinpei, Jonathan, Bo, Hongwei, and Yuxing, as well as the Gao and Valentina labs for their outstanding experimental validation of the predicted targets. This study integratesClinical Proteomic Tumor Analysis Consortium (CPTAC) proteogenomics data from 1,043 patients across 10 cancer types with additional public datasets to identify potential oncology targets. These include overexpressed and hyperactivated protein dependencies, protein dependencies associated with the loss of tumor suppressor genes, and putative neoantigens and tumor-associated antigens. These analyses, summarized at https://targets.linkedomics.org, form a comprehensive landscape of protein and peptide targets for companion diagnostics, drug repurposing, and therapy development.
[2024-08] QCB student Siyu Wang joined the group for a research rotation. Welcome, Siyu!
[2024-08] Congratulations to Yongchao on the promotion to Assistant Professor at the Breast Center!
[2024-07] John’s paper WebGestalt 2024: faster gene set analysis and new support for metabolomics and multi-omics has been published in Nucleic Acids Research. The 2024 WebGestalt update marks a major advancement, introducing support for metabolomics, streamlined multi-omics analysis capabilities, and substantial performance improvements enabled by a Rust-based implementation. Discover these updates and more at https://www.webgestalt.org.
[2024-05] CCB student Michael Lee joined our group and Dr. Cheng’s lab as a graduate student, co-mentored by Dr. Zhang and Dr. Cheng. Welcome, Michael!
[2024-04] Dr. Zhang received the Michael E. Debakey Excellence in Research Award. Congratulations!
[2024-04] Dr. Paul Shafer joined the lab as a postdoctoral research associate. Welcome, Paul!
[2024-03] Dr. Yanling Sun joined the lab as a postdoctoral research associate. Welcome, Yanling!
[2024-03] Seunghyuk, John, Lindsey, Chenwei and Bing attended the CPTAC symposium and US HUPO in Portland, OR. Congratulations to Chenwei on receiving the Meritorious Poster Award for his work on “DeepVEP: predict effects of variants on post-translational modifications with deep learning”. 
[2024-02] Breast center Spring Festival celebration. 
[2024-01] Sara’s paper Frozen tissue coring and layered histological analysis improves cell type-specific proteogenomic characterization of pancreatic adenocarcinoma has been published in Clinical Proteomics. This study demonstrates the feasibility of multi-omics data generation from tissue cores, the necessity of interval H&E stains in serial histology sections, and the utility of coring to improve analysis over bulk tissue data.
[2023-12] Xinpei’s paper Deep Learning Prediction Boosts Phosphoproteomics-Based Discoveries Through Improved Phosphopeptide Identification has been published in Molecular & Cellular Proteomics. Shotgun phosphoproteomics enables high-throughput analysis of phosphopeptides in biological samples, but low phosphopeptide identification rates in data analysis limit its potential. This paper presents DeepRescore2, a computational workflow that leverages deep learning-based predictions of retention time and fragment ion intensity to enhance phosphopeptide identification and phosphosite localization. Benchmarking against existing workflows on a synthetic phosphopeptide dataset and application to real-world biological datasets demonstrate increased sensitivity, reduced missing values, and improved insights from phosphoproteomics-based biological analyses. DeepRescore2 is available at https://github.com/bzhanglab/DeepRescore2.
[2023-12] Holiday bowling party. 
[2023-12] Dr. Seunghyuk Choi joined the lab as a postdoctoral research associate. Welcome, Seunghyuk!
[2023-11] Dr. Wenrong Chen joined the lab as a postdoctoral research associate. Welcome, Wenrong!
[2023-11] Sara’s paper IDPpub: Illuminating the Dark Phosphoproteome Through PubMed Mining has been published in Molecular & Cellular Proteomics. Phosphorylation is an essential component of cellular signaling, and phosphoproteomics enables global identification and quantification of phosphosites from biological samples. However, interpretation of phosphoproteomic findings is hindered by our limited knowledge on functions, phenotype associations, and regulating enzymes of the phosphosites. We developed a computational pipeline that uses BioBERT to extract phosphorylation sites from biomedical abstracts. The pipeline further aligns the sites to human and mouse reference sequences to facilitate computational applications and intersection with mass spectrometry experiments. The extracted evidence sentences can be used to identify regulating enzymes and biological functions. We made all data available in the IDPpub web portal for easy exploration.
[2023-10] The review article Current perspectives on mass spectrometry-based immunopeptidomics: the computational angle to tumor antigen discovery co-authored by Drs. Zhang and Bassani-Sternberg has been published in The Journal for ImmunoTherapy of Cancer.
[2023-10] The paper ClinicalOmicsDB: exploring molecular associations of oncology drug responses in clinical trials led by James and John has been published in Nucleic Acids Research. Congratulations! This paper describes ClinicalOmicsDB, a web application for exploring molecular associations of oncology drug responses in clinical trials. This database encompasses data from 40 clinical trial studies and a total of 5913 patients, including 1224 patients treated with immunotherapy. Three case studies were presented to demonstrate the utility of this resource in human cancer research.
[2023-10] QCB student Minhang Xu joined the group for a research rotation. Welcome, Minhang!
[2023-09] Yongchao’s paper SEPepQuant enhances the detection of possible isoform regulations in shotgun proteomics has been published in Nature Communications. Shotgun proteomics is crucial for identifying and quantifying proteins in biomedical research. However, characterizing protein isoforms is challenging due to shared peptides among proteins. We introduce SEPepQuant, a graph theory-based method to tackle this challenge. SEPepQuant addresses limitations of existing methods, enhancing isoform characterization, identifying isoform-level regulation events, and facilitating cross-study comparisons. Our results support a significant role of protein isoform regulation in normal and disease processes, making SEPepQuant valuable for biological and translational research. Source code is available in the Zhang Lab GitHub.
[2023-09] Jonathan attended the HUPO Conference held in Busan, Korea, where he delivered an oral presentation titled “Pan-cancer proteogenomics expands the landscape of therapeutic targets”. He was honored with a travel award in recognition of this work. Congratulations! 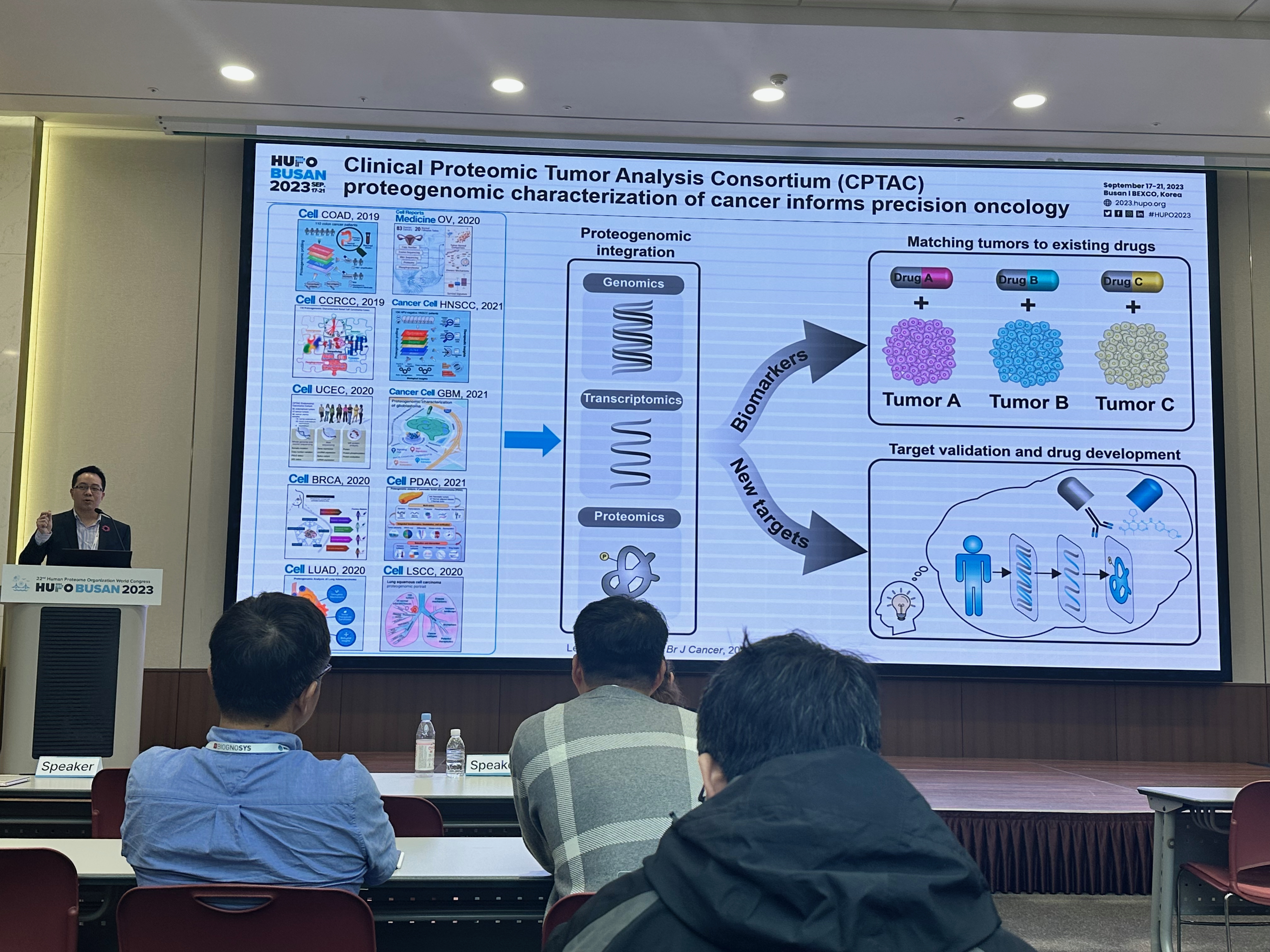
[2023-08] Our paper A proteogenomics data-driven knowledge base of human cancer has been published in Cell Systems. Congratulations to Yuxing, Sara, and all co-authors! This paper describes LinkedOmicsKB, a knowledge base built upon consistently processed and systematically precomputed CPTAC pan-cancer proteogenomics data. With approximately 40,000 gene-, protein-, mutation-, and phenotype-centric web pages, it enables anyone with internet access to conduct meaningful inquiries into CPTAC data, facilitating data-driven scientific discoveries. The paper uses three case studies to illustrate the practical utility of LinkedOmicsKB in providing new insights into genes, phosphorylation sites, somatic mutations, and cancer phenotypes.
[2023-08] The CPTAC perspective article Proteogenomic data and resources for pan-cancer analysis has been published in Cancer Cell. Congratulations to Yongchao and all co-authors! This article describes efforts by the CPTAC pan-cancer working group in data harmonization, data dissemination, and the provision of computational resources to facilitate biological discoveries. All processed data tables can be accessed at the Proteome Data Commons.
[2023-08] The CPTAC study Proteogenomic insights suggest druggable pathways in endometrial carcinoma has been published in Cancer Cell. Congratulations to Yongchao and all co-authors! Some of the key findings include identifying two peptides that can predict antigen processing and presentation machinery activity, revealing a potential role for metformin treatment in non-diabetic patients with elevated MYC activity, discoverying PIK3R1 in-frame indels as a primary driver of elevated AKT phosphorylation and increased sensitivity to AKT inhibitors, and connecting CTNNB1 hotspot mutations to pS45 phosphorylation-induced degradation of β-catenin.
[2023-08] QCB student Juliana Yue joined the group for a research rotation. Welcome, Juliana!
[2023-06] Most of our lab members participated in the ASMS Conference held in Houston, where we showcased our recent work. One notable highlight was Yongchao delivering an oral presentation titled “SEPepQuant Enhances the Detection of Possible Isoform Regulations in Shotgun Proteomics”. 
[2023-05] QCB student Tobie Lee joined the group for a research rotation. Welcome, Tobie!
[2023-05] Faye successfully defended her PhD thesis, congratulations, Dr. Jiang! She will be joining AstraZeneca as a Senior Computational Biologist next month. Best wishes for a bright future!

[2023-04] Faye, James, Jonathan, Sara and Dr. Zhang attended the AACR Conference in Orlando, FL. James received a 2023 AACR-Sanofi Scholar-in-Training Award for his work on “Bridging the gap between clinical-omics and machine learning to improve cancer treatment”, and Johnathan received the same award for the work on “Pan-cancer proteogenomics expands the landscape of therapeutic targets”. Congratulations! Both works were also selected for oral presentations. Faye did a poster presentation on CoPheeMap, and Sara did a poster presentation on LinkedOmicsKB.

[2023-04] Bo’s paper PepQuery2 democratizes public MS proteomics data for rapid peptide searching has been published in Nature Communications. One of the most important milestones in proteomics is the Amsterdam Principles, which require mandatory raw MS/MS data deposition to promote broad reuse of the data. However, because of the challenges involved in understanding, downloading, analyzing, and interpreting MS/MS data, investigation and reuse of these public data are largely restricted to computational proteomics researchers. By enabling rapid identification of any known or novel peptide sequences of interest in any local or publicly available MS-based proteomics datasets in a targeted manner, PepQuery2 provides a practical solution that makes public MS/MS data easily useful to the general research community. Both the command line version and the web version of PepQuery2 are available at http://www.pepquery.org. The source code of PepQuery2 is available at https://github.com/bzhanglab/PepQuery.
[2023-03] Congratulations to Lindsey for passing the CCB PhD Qualifying Exam!
[2023-03] Faye, Lindsey and Dr. Zhang attended the US HUPO Conference in Chicago. Faye received a Travel Award for her work on “Illuminating the Dark Cancer Phosphoproteome Through a Machine Learned Co-Regulation Map of 30,000 Phosphosites”, which was also selected for an oral presentation. Dr. Zhang received the Gilbert S. Omenn Computational Proteomics Award and gave an award talk entitled “Embracing Complexity, Seeking Simplicity”. Congratulations!

[2023-01] Lunch party to celebrate the new year and welcome new lab members!

[2023-01] Duy Pham and John Elizarraras joined the group as Bioinformatics Programmers. Welcome, Duy and John!
[2023-01] CCB student Evelyn de Groot joined the group for a research rotation. Welcome, Evelyn!
[2022-12] Lindsey  gave an oral presentation on “Tripartite graph modeling enables comprehensive protein isoform characterization in shotgun proteomics” at the HUPO 2022 conference in Cancun.
gave an oral presentation on “Tripartite graph modeling enables comprehensive protein isoform characterization in shotgun proteomics” at the HUPO 2022 conference in Cancun.
[2022-11] Faye, James, and Lindsey were selected for oral presentation at the 17th Annual Breast Center Retreat, and James won the 2nd place prize for his presentation entitled “ClinicalOmicsDB – Bridging the gap between clinical omics data and machine learning”. Congratulations!
[2022-10] QCB students Jiaye Chen and Daniel Palacios joined the group for a research rotation. Welcome, Jiaye and Daniel!
[2022-08] Dr. Chenwei Wang joined the lab as a postdoctoral research associate. Welcome, Chenwei!
[2022-08] QCB student Xuqian Tan joined the group for a research rotation. Welcome, Xuqian!
[2022-07] Our U01 application entitled “Illuminating understudied druggable proteins using pan-cancer proteogenomics data” has been selected for funding by the Illuminating the Druggable Genome (IDG) consortium.
[2022-07] James was offered a position in the CTR Certificate of Added Qualification program and the program’s T32 grant. The CAQ training is designed to develop leaders in translational research who are well equipped to translate discoveries from the laboratory to the clinic to the benefit of human health. Congratulations!
[2022-06] The Office of Cancer Clinical Proteomics Research at the National Cancer Institute (NCI) has reaffirmed its commitment to furthering proteogenomics research by announcing the next round of Clinical Proteomic Tumor Analysis Consortium (CPTAC) centers. As part of this new phase, our lab will continue to serve as a Proteogenomic Data Analysis Center (PGDAC) over the next five years.
[2022-06] Xinpei and Sara gave oral presentations about their computational tools DeepRescore2 and IDPpub, respectively, at the 70th ASMS Conference on Mass Spectrometry and Allied Topics (ASMS 2022) took place 5-9 June 2022 in Minneapolis, MN, USA. DeepRescore2 leverages deep learning to improve phosphopeptide identification and site localization in phosphoproteomics, whereas IDPpub aims to illuminate the dark phosphoproteome through PubMed mining.
[2022-06] Byron Jia, a rising senior at Carleton College joined the group for a summer internship through the SMART program. Welcome, Byron!
[2022-05] CCB student Lindsey Olsen joined the group as a graduate student. Welcome, Lindsey!
[2022-03] Faye has been awarded funding on the CPRIT BCM Comprehensive Cancer Training Program to support her research on “Leveraging Artificial Intelligence to Illuminate the Cancer Phosphoproteome”. Congratulations!
[2022-02] Dr. Zhang is a recipient of a CPRIT Individual Investigator Research Award for Computational Systems Biology.
[2022-01] Congratulations to James for passing the QCB PhD Qualifying Exam!
[2021-12] We had a much needed and deserved Holiday Party at the Hermann Park. Great food and great fun! Special thanks to James for putting this superb event together!


[2021-11] Faye’s paper Deep learning-derived evaluation metrics enable effective benchmarking of computational tools for phosphopeptide identification has been published in Molecular & Cellular Proteomics. The benchmark metrics demonstrated in this study will enable users to select computational pipelines and parameters for routine analysis of phosphoproteomics data and will offer guidance for developers to improve computational methods. Congratulations to Faye!
[2021-10] Congratulations to Jonathan on winning the 2nd place prize for his oral presentation at the 16th Annual Breast Center Retreat!
[2021-09] Xinpei’s paper caAtlas: An immunopeptidome atlas of human cancer has been published in iScience. This paper reports an immunopeptidome atlas of human cancer constructed through an extensive collection of 43 published immunopeptidomic datasets and standardized analysis of 81.6 million MS/MS spectra using Open-pFind. The study greatly expands the current knowledge of MHC-bound antigens, including an unprecedented characterization of post-translationally modified antigens. Further analysis of these data provides evidence for tumor specific-presentation of post translationally modified antigens, cancer testis antigens and cancer type-specific tumor associated antigens. All these data together with annotated MS/MS spectra supporting identification of each antigen are available in an easily browsable web portal named cancer antigen atlas (caAtlas). caAtlas provides a central resource for the selection and prioritization of MHC-bound peptides for in vitro HLA binding assay and immunogenicity testing, which will pave the way to eventual development of cancer immunotherapies. Congratulations to Xinpei, Yuxing, and all coauthors!
[2021-09] The CPTAC study Proteogenomic characterization of pancreatic ductal adenocarcinoma has been published in Cell. Congratulations to Chen and all co-authors!
[2021-08] The CPTAC study A proteogenomic portrait of lung squamous cell carcinoma has been published in Cell. Congratulations to Sara and all co-authors!
[2021-06] Kuan Huang joined the lab as a postdoctoral research associate. Welcome, Kuan!
[2021-05] The paper describing the crowdsourcing precisionFDA NCI-CPTAC Multi-omics Enabled Sample Mislabeling Correction Challenge has been published in Patterns. Congratulations to Seungyeul, Zhiao, Bo, SoonJye, and all co-authors! The final collaborative product, COSMO, can be accessed at https://github.com/bzhanglab/COSMO.
[2021-04] Zhiao’s paper Feature selection methods for protein biomarker discovery from proteomics or multi-omics data has been published in Molecular & Cellular Proteomics. The algorithms ProMS and ProMS_mo show good performance, enable functional interpretation of the identified markers, and provide alternative choices for each identified marker to facilitate a robust transition to the verification and validation platforms. The software can be downloaded at https://github.com/bzhanglab/proms.
[2021-04] Xinpei received the AACR Women in Cancer Research (WICR) Scholar Award for her work on caAtlas, which was presented at the AACR 2021 conference. Meanwhile, Jonathan received an AACR Doreen J. Putrah Cancer Research Foundation Scholar-in-Training Award recognizing his work on chemotherapy response and potential therapeutic targets to overcome resistance in triple-negative breast cancer. Congratulations!
[2021-03] QCB student Chang In Moon (James) joined the group as a graduate student. Welcome, James!
[2021-03] Xinpei has been selected for the trainee award by the US HUPO 2021 conference committee for her work on caAtlas and will give an award talk at the conference. Meanwhile, Faye has received honorable mention for her work on “Deep learning-derived evaluation metrics for benchmarking computational pipelines for the analysis of large-scale phosphoproteomic datasets”, which has also been selected for an oral presentation. Congratulations!
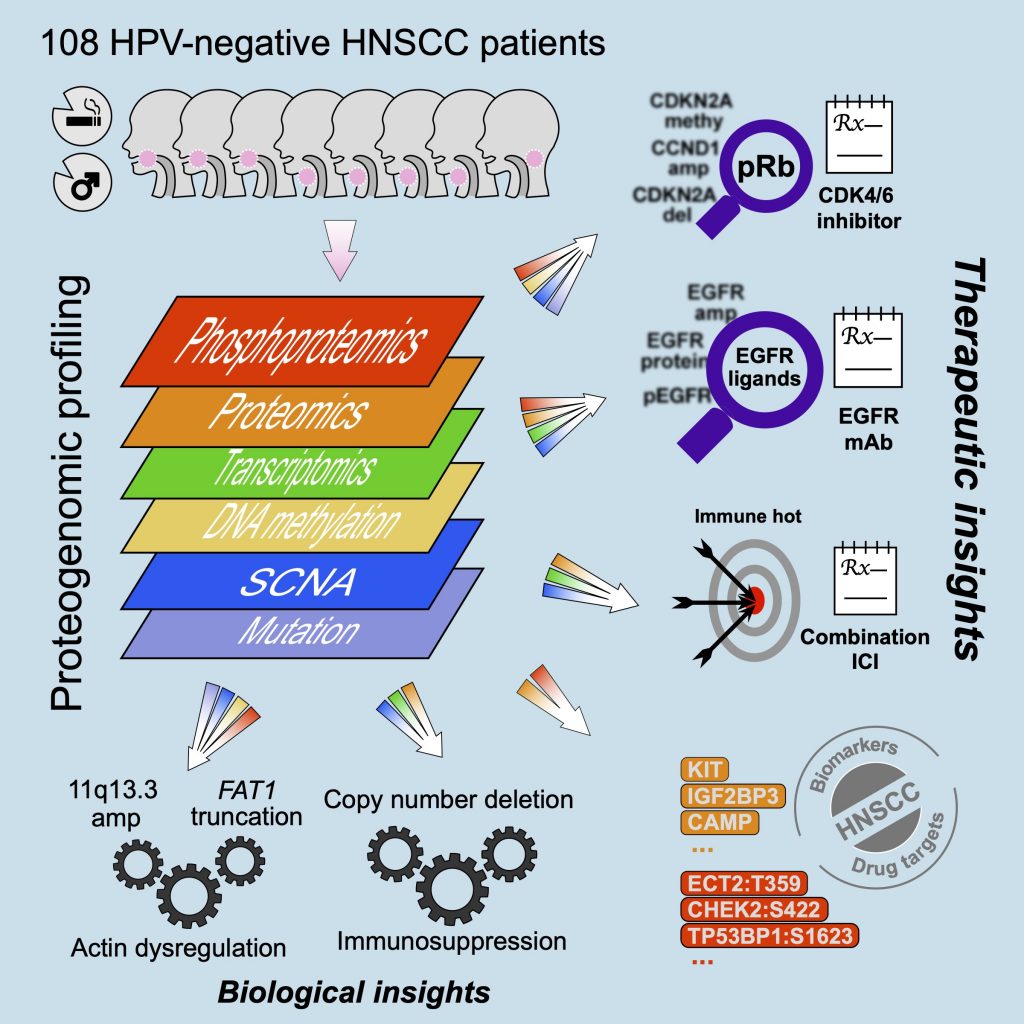
[2021-01] The CPTAC study Proteogenomic insights into the biology and treatment of HPV-negative head and neck squamous cell carcinoma has been published in Cancer Cell. Congratulations to Chen, Sara, and all co-authors! In addition to providing more complete biological understanding of HPV-negative HNSCC, this study demonstrates the potential of proteogenomics as a therapeutic hypothesis generator. First, focused characterization of the target proteins and pathways of the standard-of-care or investigational drugs identifies biomarkers that may help match HNSCC patients to available treatments. Moreover, unbiased exploratory analysis of proteogenomic data further reveals new putative therapeutic targets for further experimental validation.
[2021-01] Diyahir Campos joined the lab as a bioinformatics programmer. Welcome, Diyahir!
[2020-11] The Proteomics special issue, “Computational Proteomics: Focus on Deep Learning“, is now online. This special issue, edited by Bo and Dr. Zhang, brings together 17 original research, review, and perspective articles on applying novel computational technologies, with a focus on deep learning methods, to the analysis of MS-based proteomics data.

[2020-10] Jiayi Luo from the CCB program joined the lab for a research rotation. Welcome, Jiayi!
[2020-10] James Moon from the QCB program joined the lab for a research rotation. Welcome, James!
[2020-09] A review article led by Bo, Deep learning in proteomics, has been published in Proteomics. This paper provides a comprehensive overview of deep learning applications in proteomics including retention time prediction, MS/MS spectrum prediction, de novo peptide sequencing, PTM prediction, major histocompatibility complex-peptide binding affinity prediction, and protein structure prediction. A list of the applications described in the paper can be found at our GitHub site.
[2020-08] Kai’s paper DeepRescore: leveraging deep learning to improve peptide identification in immunopeptidomics has been published in Proteomics. Congratulations, Kai! DeepRescore integrates peptide features derived from deep learning predictions, namely accurate retention time and MS/MS spectra predictions, into the rescoring of peptide-spectrum matches, which increases both the sensitivity and reliability of the identification of MHC-bound peptides and neoantigens. DeepRescore is developed using NextFlow and Docker and is available at https://github.com/bzhanglab/DeepRescore.
[2020-08] Congratulations to Michael on successfully defending his PhD thesis!
[2020-08] Mariah Berner from the CCB program joined the lab for a research rotation. Welcome, Mariah!
[2020-07] Sara’s review article Using phosphoproteomics data to understand cellular signaling: a comprehensive guide to bioinformatics resources has been published in Clinical Proteomics.
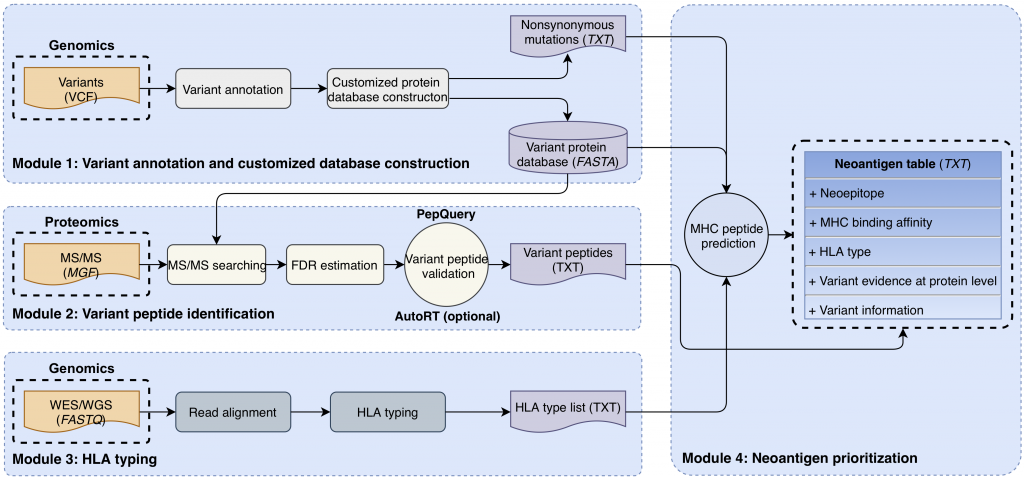 [2020-04] Bo’s paper Cancer neoantigen prioritization through sensitive and reliable proteogenomics analysis has been published in Nature Communications. Congratulations, Bo! Identifying mutation-derived neoantigens by proteogenomics requires robust strategies for quality control. In this paper, we propose peptide retention time as an evaluation metric for proteogenomics quality control methods, and develop a deep learning algorithm AutoRT for accurate retention time prediction. Our systematic evaluation, using the proposed retention time metric, provides insights and practical guidance on the selection of quality control strategies. We implement the recommended strategy in a computational workflow named NeoFlow to support proteogenomics-based neoantigen prioritization, enabling more sensitive discovery of putative neoantigens.
[2020-04] Bo’s paper Cancer neoantigen prioritization through sensitive and reliable proteogenomics analysis has been published in Nature Communications. Congratulations, Bo! Identifying mutation-derived neoantigens by proteogenomics requires robust strategies for quality control. In this paper, we propose peptide retention time as an evaluation metric for proteogenomics quality control methods, and develop a deep learning algorithm AutoRT for accurate retention time prediction. Our systematic evaluation, using the proposed retention time metric, provides insights and practical guidance on the selection of quality control strategies. We implement the recommended strategy in a computational workflow named NeoFlow to support proteogenomics-based neoantigen prioritization, enabling more sensitive discovery of putative neoantigens.
[2020-02] The CPTAC study Proteogenomic Characterization of Endometrial Carcinoma has been published in Cell. Congratulations to Yongchao and all co-authors!
[2020-01] Our collaborative study with Drs. Ellis, Carr and other colleagues entitled Microscaled proteogenomic methods for precision oncology has been published in Nature Communications. Congratulations to Eric and all co-authors!
[2019-12] Congratulations to Faye for passing the QCB PhD Qualifying Exam!
[2019-10] Chaozhong Liu from the QCB program joined the lab for a research rotation. Welcome, Chaozhong!
[2019-07] Zifan Zhao from the Cancer & Cell Biology program joined the lab for a research rotation. Welcome, Zifan!
[2019-07] Linhua Wang from the QCB program joined the group for a research rotation. Welcome, Linhua!
[2019-05] Sara and Zhiao’s paper Graph algorithms for condensing and consolidating gene set analysis results has been published in Molecular & Cellular Proteomics. Two graph algorithms were used to integrate gene set analysis results from multiple experiments, such as multi-omics or pan-cancer studies. Specifically, a weighted set cover algorithm was used to reduce redundancy of gene sets identified in a single experiment, and then affinity propagation was used to consolidate similar gene sets identified from multiple experiments into clusters and to automatically determine the most representative gene set for each cluster. This has been implemented in an R package Sumer, which is available at https://github.com/bzhanglab/sumer.
[2019-05] Yuxing’s paper WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs has been published in Nucleic Acids Research. There are five major changes in this new version: 1) We have completely redesigned result visualizations and user interfaces to improve user-friendliness and to provide multiple types of interactive and publication-ready figures. 2) To address the growing and unique need for phosphoproteomics data interpretation, we have implemented phosphosite set analysis to identify important kinases from phosphoproteomics data. 3) To facilitate comprehension of the enrichment results, we have implemented two methods to reduce redundancy between enriched gene sets. 4) We introduced a web API for other applications to get data programmatically from the WebGestalt server or pass data to WebGestalt for analysis. 5) We wrapped the core computation into an R package called WebGestaltR for users to perform analysis locally or in third party workflows.
[2019-05] QCB student Jiasheng Wang joined the group as a graduate student. Welcome, Jiasheng!
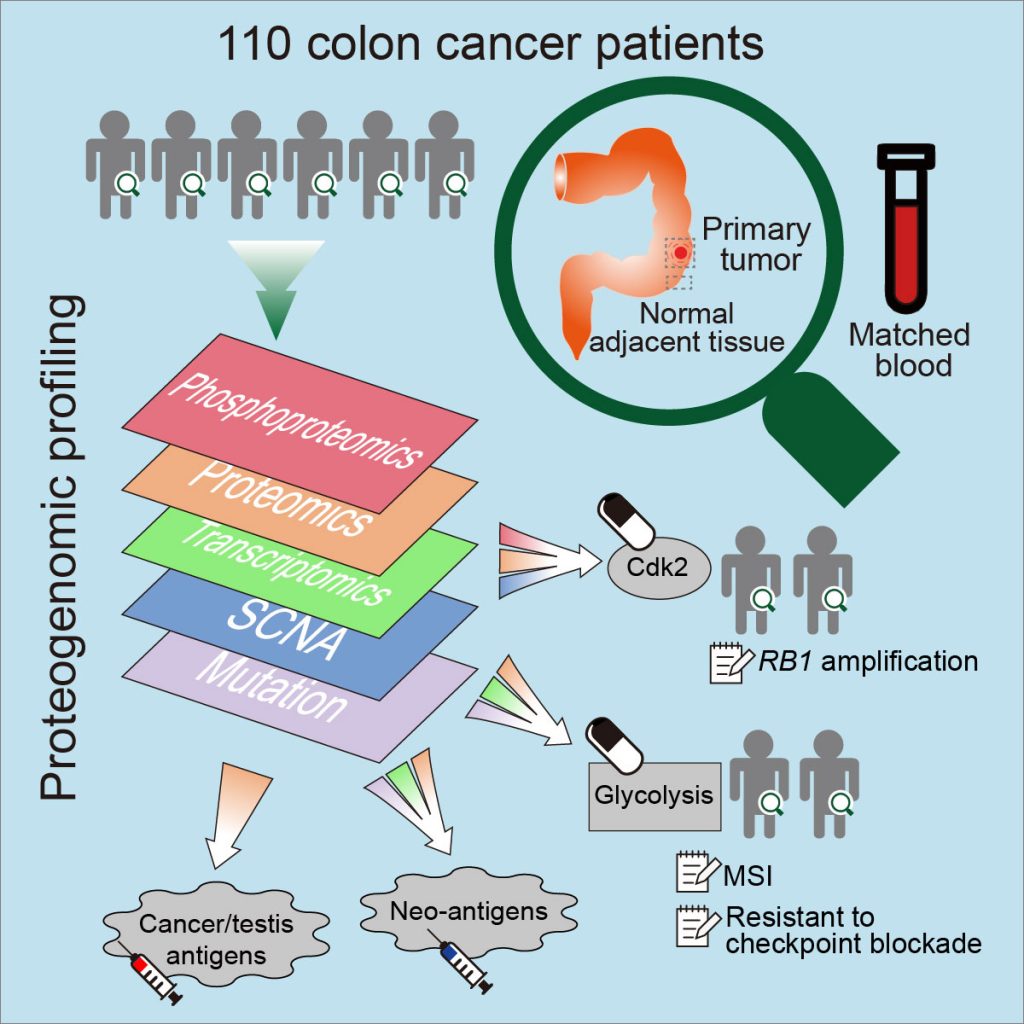 [2019-05] The CPTAC study Proteogenomic Analysis of Human Colon Cancer Reveals New Therapeutic Opportunities has been published in Cell. This is an extension of the 2014 CPTAC colorectal cancer study. In addition to confirming the value of proteogenomic integration in uncovering novel cancer biology, this new study further demonstrated the utility of proteogenomics in therapeutic hypothesis generation. For example, phosphoproteomics data associated Rb phosphorylation with increased proliferation and decreased apoptosis in colon cancer, which explains why this classical tumor suppressor is amplified in colon tumors and suggests a rationale for targeting Rb phosphorylation in colon cancer. As another example, proteomics identified an association between decreased CD8 T cell infiltration and increased glycolysis in microsatellite instability-high (MSI-H) tumors, suggesting glycolysis as a potential target to overcome the resistance of MSI-H tumors to immune checkpoint blockade. The primary and processed datasets are available in publicly accessible data repositories and portals (e.g., LinkedOmics), which we hope will allow new biological discoveries and therapeutic hypothesis generation.
[2019-05] The CPTAC study Proteogenomic Analysis of Human Colon Cancer Reveals New Therapeutic Opportunities has been published in Cell. This is an extension of the 2014 CPTAC colorectal cancer study. In addition to confirming the value of proteogenomic integration in uncovering novel cancer biology, this new study further demonstrated the utility of proteogenomics in therapeutic hypothesis generation. For example, phosphoproteomics data associated Rb phosphorylation with increased proliferation and decreased apoptosis in colon cancer, which explains why this classical tumor suppressor is amplified in colon tumors and suggests a rationale for targeting Rb phosphorylation in colon cancer. As another example, proteomics identified an association between decreased CD8 T cell infiltration and increased glycolysis in microsatellite instability-high (MSI-H) tumors, suggesting glycolysis as a potential target to overcome the resistance of MSI-H tumors to immune checkpoint blockade. The primary and processed datasets are available in publicly accessible data repositories and portals (e.g., LinkedOmics), which we hope will allow new biological discoveries and therapeutic hypothesis generation.
[2019-04] QCB student Wen Jiang (Faye) joined the group as a graduate student. Welcome, Faye!
[2019-03] Dr. Xinpei Yi joined the lab as a postdoctoral research fellow. Xinpei just graduated from the Institute of Applied Mathematics, Chinese Academy of Sciences, Beijing, with a PhD degree in Probability Theory and Mathematical Statistics and Bioinformatics. Welcome, Xinpei!
[2019-01] Bo’s paper PepQuery enables fast, accurate, and convenient proteomic validation of novel genomic alterations has been published in Genome Research. Congratulations, Bo! PepQuery is a peptide-centric search engine that allows quick and easy proteomic validation of genomic alterations without customized database construction. Next generation sequencing-based genomic studies continuously identify new genomic alterations that may lead to novel protein sequences, which are attractive candidates for disease biomarkers and therapeutic targets after proteomic validation. The popular approach for proteomic validation requires customized database construction and a full evaluation of all possible spectrum-peptide pairs, which is time-consuming. We implemented PepQuery as both stand-alone and web-based applications. The stand-alone version supports batch analysis and user-provided MS/MS data. The web version provides access to more than half a billion MS/MS spectra from the Clinical Proteomic Tumor Analysis Consortium (CPTAC) and other cancer proteomic studies, making MS/MS data directly available and useful to scientists outside the proteomics community. PepQuery can be accessed at http://www.pepquery.org.
[2019-01] Dr. Jonathan Lei joined the lab as a postdoctoral research fellow supported by the Breast Center T32 training program. Jonathan just got his PhD in Translational Biology & Molecular Medicine from BCM. Welcome, Jonathan!
[2019-01] Ahmed Gad from TBMM interdisciplinary program joined the lab for a research rotation. Welcome, Ahmed!
[2019-01] Jiasheng Wang from the QCB program joined the group for a research rotation. Welcome, Jiasheng!
[2018-12] Kai Li joined the lab as a bioinformatics programmer. Welcome, Kai!
[2018-10] Wen Jiang (Faye) from the QCB program joined the group for a research rotation. Welcome, Faye!
[2018-09] Bo’s paper PDV: an integrative proteomics data viewer has been published in Bioinformatics. Congratulations, Bo! PDV is a lightweight visualization tool that enables intuitive and fast exploration of diverse, large-scale proteomics datasets on standard desktop computers in both graphical user interface and command line modes.The software and the user manual are freely available at http://pdv.zhang-lab.org . The source code is available at https://github.com/wenbostar/PDV.
[2018-07] Congratulations to Sara on successfully defending her Master’s thesis!
[2018-05] Four students joined the group for summer research experience. Damilola Omotajo is an incoming graduate student in the MHG program; she has a B.S. degree in Mathematics with Minor in Biology and an M.S. degree in Biology with Minor in Statistical Methods from the Sam Houston State University.Paul Jiang, a rising junior at the Rice University pursuing a degree in Computer Science, joined the group as a Research Observer. Both Aaron Westbrook and Kailey Ferger are supported by the SMART program. Aaron just graduated from the University of St. Thomas with a B.S. degree in Bioinformatics, and this is his second summer in our group. Kailey is a rising senior at the University of Rochester pursuing a degree in Computational Biology. Welcome our new members!
[2018-02] Mr. Ram Srinivasan joined the group as a Senior Bioinformatics Programmer. Ram received his B.S. degree in Bioinformatics from the SASTRA University in India and M.S. degree in Bioinformatics from the NYU Polytechnic School of Engineering. He worked at the Icahn School of Medicine at Mount Sinai for three years before joining our lab. Welcome, Ram!
[2018-02] Dr. Yuxing Liao joined the group as a Programmer/Analyst II. Yuxing received his B.S. degree in Biological Science from the University of Science and Technology in China and just completed his Ph.D. study on Molecular Biophysics with Computational and Systems Biology specialty at the University of Texas Southwestern Medical Center. Welcome, Yuxing!
[2018-02] Mr. Noel Namai joined the group as a Programmer/Analyst I. Noel received his M.S. degree in Bioinformatics from the Northeastern University and B.S. degree in Mathematics from the Maseno University in Kenya. He worked at the Seven Bridges Genomics for two years before joining us. Welcome, Noel!
[2018-01] Mr. Leo Zhang joined the group as a Research Technician. Leo received his B.S. degree in Biological Science from the Colorado State University with a Minor in Mathematics. Welcome, Leo!
[2017-12] Xiaojing’s paper Detection of proteome diversity resulted from alternative splicing is limited by trypsin cleavage specificity. has been published in Molecular & Cellular Proteomics. Congratulations, Xiao Jing! Alternative splicing dramatically increases transcriptome complexity but its contribution to proteome diversity remains controversial. Exon-exon junction spanning peptides provide direct evidence for the translation of specific splice isoforms and are critical for delineating protein isoform complexity. In this paper, we studied the proteomic coverage of exon-exon junctions in three publicly available proteomics data sets and found that trypsin preferentially cleaves exon-exon junctions and thus hinders the detection of junction-spanning peptides. This phenomenon was explained by evolutionarily conserved preferential nucleotide usage at exon boundaries according to nucleotide sequence analysis of five eukaryotic genomes. Our in silico and experimental analyses showed that complementary digestion schemes are essential to study the translation of alternative mRNA splicing to proteome diversity.
[2017-11] Dr. Eric Jaehnig joined the group as a Research Associate. Eric received his Ph.D. degree in Biology from the University of California, San Francisco, followed by postdoctoral training in systems biology at the Ludwig Institute for Cancer Research. To enhance his quantitative skills, Eric recently completed a Master’s degree in Bioengineering from the Rice University. Welcome, Eric!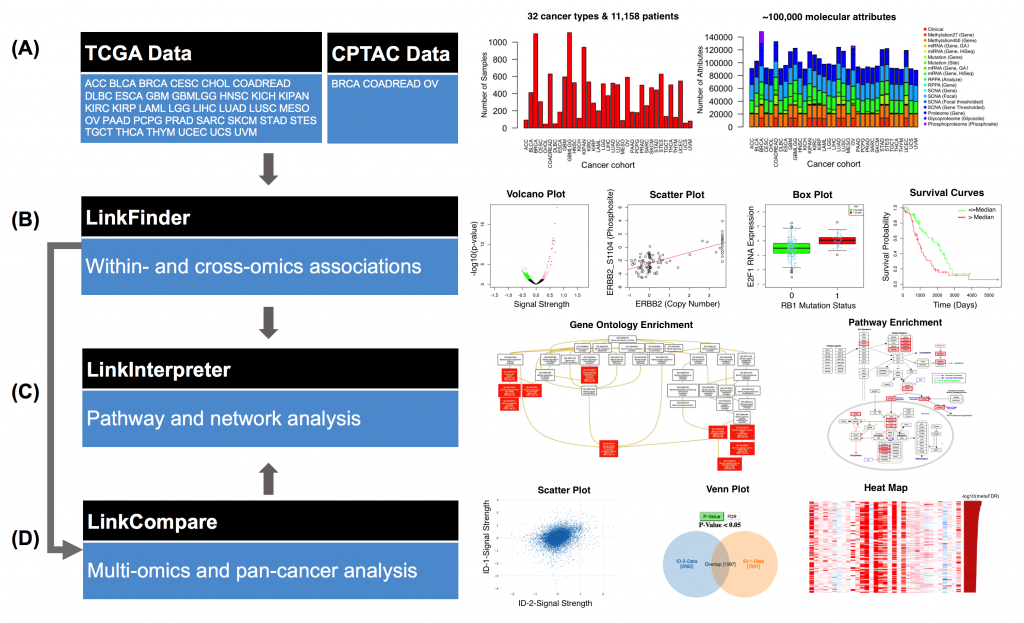
[2017-11] Suhas’ paper LinkedOmics: analyzing multi-omics data within and across 32 cancer types has been published in Nucleic Acids Research. LinkedOmics is a new and unique resource for disseminating and analyzing The Cancer Genome Atlas (TCGA) data. It is also the first multi-omics database that integrates mass spectrometry (MS)-based global proteomics data generated by the Clinical Proteomic Tumor Analysis Consortium (CPTAC) on selected TCGA tumor samples. LinkedOmics has three analysis modules. The LinkFinder module allows flexible exploration of associations between a molecular or clinical attribute of interest and all other attributes, providing the opportunity to analyze and visualize associations between billions of attribute pairs for each cancer cohort. The LinkCompare module enables easy comparison of the associations identified by LinkFinder, which is particularly useful in multi-omics and pan-cancer analyses. The LinkInterpreter module transforms identified associations into biological understanding through pathway and network analysis. In this paper, we used five case studies to demonstrate that LinkedOmics provides a unique platform for biologists and clinicians to access, analyze and compare cancer multi-omics data within and across tumor types. Although the current version of LinkedOmics includes only TCGA and CPTAC data, it can be easily extended to support other cohort-based multi-omics studies.
[2017-10] Dr. Hyokyeong Lee joined the group as a Research Associate. Hyokyeong received her Ph.D. degree in Computer Science from the University of Southern California and was a Postdoctoral Associate in the Department of Computational and Systems Biology at University of Pittsburgh. Welcome to the lab, HyoKyeong!
[2017-10] Chaohao Gu from the SCBMB program joined the group for a research rotation. Welcome, Chaohao!
[2017-08] PepQuery, a peptide-centric search engine for novel peptide identification and validation, is now online.
[2017-08] Jingyuan Hu from the SCBMB program joined the group for a research rotation. Welcome, Jingyuan!
[2017-07] Charindu Ranmuthu, a rising junior from the Queens’ College, Cambridge joined the group for a summer internship. Welcome, Cha!
[2017-06] Our collaborative study with Drs. Sieber, Liebler, Burgess, Chen and their group members on the integrative proteogenomic analysis of colorectal cancer cell lines and human tumors has been published in Gastroenterology in a paper entitled Colorectal cancer cell line proteomes are representative of primary tumors and predict drug sensitivity.
[2017-05] Aaron Westbrook, a rising senior at the University of St. Thomas in Houston joined the group for a summer internship through the SMART program. Welcome, Aaron!
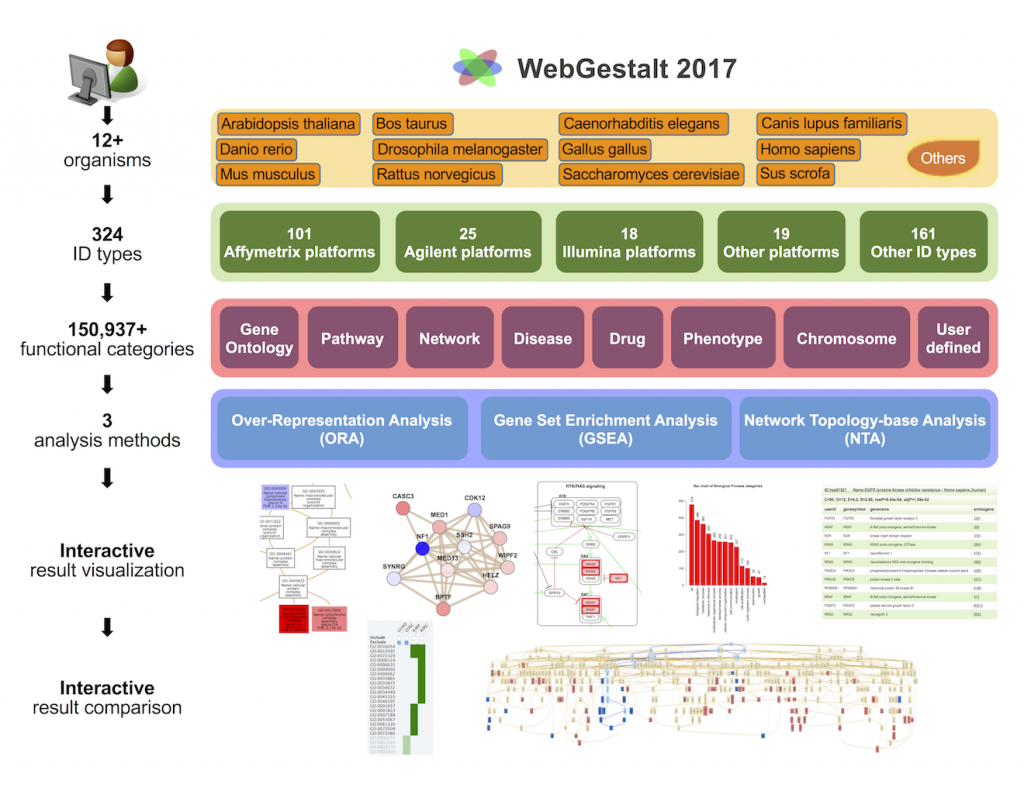 [2017-05] Jing’s paper WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit has been published in Nucleic Acids Research. The WebGestalt 2017 knowledge base supports 12 organisms, 324 gene identifiers from various databases and technology platforms, and 150,937 functional categories from public databases and computational analyses. Importantly, omics data with gene identifiers not supported by WebGestalt and functional categories not included in the WebGestalt database can also be uploaded for enrichment analysis. The analysis module supports Over-Representation Analysis, Gene Set Enrichment Analysis, and Network Topology-based Analysis. The input interface is simplified, the output interface allows interactive and efficient exploration, and the complete set of results can be easily downloaded. The new GOView tool makes it easy to compare multiple sets of GO enrichment results in the context of the GO directed acyclic graph (DAG).
[2017-05] Jing’s paper WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit has been published in Nucleic Acids Research. The WebGestalt 2017 knowledge base supports 12 organisms, 324 gene identifiers from various databases and technology platforms, and 150,937 functional categories from public databases and computational analyses. Importantly, omics data with gene identifiers not supported by WebGestalt and functional categories not included in the WebGestalt database can also be uploaded for enrichment analysis. The analysis module supports Over-Representation Analysis, Gene Set Enrichment Analysis, and Network Topology-based Analysis. The input interface is simplified, the output interface allows interactive and efficient exploration, and the complete set of results can be easily downloaded. The new GOView tool makes it easy to compare multiple sets of GO enrichment results in the context of the GO directed acyclic graph (DAG).
[2017-02] Dr. Chen Huang joined the group as a Postdoctoral Research Fellow. Chen just received his Ph.D. degree in Biomedical Science from the Vanderbilt University earlier this month. Welcome to the lab, Chen!
[2017-02] Mr. Bo Wen joined the group as a Bioinformatics Programmer. Bo received his Bachelor’s degree in Bioinformation Technology from the Huazhong University of Science and Technology in China in 2010. After graduation, he has been working in the Proteomics Research Group at the BGI-Shenzhen, where he has developed and published multiple bioinformatics tools for proteomics and proteogenomics. Welcome, Bo!
[2016-11] Yongchao’s paper Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes has been published in Scientific Reports. Congratulations, Yongchao!
[2016-11] From GenomeWeb.com: Proteomic Data Improves Gene Function Predictions, CPTAC Team Finds.
[2016-11] Jing’s paper Proteome profiling outperforms transcriptome profiling for co-expression based gene function prediction has been published in Molecular & Cellular Proteomics. Congratulations, Jing!
[2016-11] Zhang Lab welcomes visiting scholar Professor Sang-Won Lee from the Korea University and his student Jeong Eun So (Jessie). Sang-Won and Jessie will be staying with us until January 2017.
[2016-10] Baylor welcomes new McNair Scholar, Dr. Bing Zhang
[2016-08] Zhang Lab has moved from VUMC to BCM.
[2016-07] Congratulations to Matt on successfully defending his Master’s thesis!
[2016-06] Congratulations to Jason on successfully defending his Master’s thesis!
[2015-12] Qi’s paper Integrative omics analysis reveals post-transcriptionally enhanced protective host response in colorectal cancers with microsatellite instability has been published in the Journal of Proteome Research. The paper introduces a resampling-based regression method that integrates transcriptomic and proteomic datasets to accurately identify genes subject to differential post-transcriptional regulation. Applying the method to the TCGA colorectal tumors revealed post-transcriptionally innitiated protective host response in cancers with microsatellite instability.
[2015-12] Xiaojing’s paper proBAMsuite, a bioinformatics framework for genome-based representation and analysis of proteomics data has been published in Molecular & Cellular Proteomics(MCP). This paper introduces protein BAM (proBAM), a new file format for organizing peptide spectrum matches (PSMs) within the context of the genome. The proBAMsuite includes two R packages, proBAMr and proBAMtools, for generating and analyzing proBAM files, respectively. The study demonstrated the utility of the proBAMsuite in facilitating efficient genome-based sharing, interpretation and integration of proteomics and proteogenomics data.
[2015-12] Matt received the 2015 DBMI Research Staff Excellence Award. Congratulations!
[2015-12] Junhui Shen M.S. joined the group as a visiting scholar. Junhui is an Associate Professor at the Information Center of the Beijing University of Chinese Medicine. Welcome to the group!
[2015-10] QCB student Mary Benton joined the group for a research rotation. Welcome, Mary!
[2015-08] MSTP student Patrick Wu joined the group for a research rotation. Welcome, Patrick!
[2015-07] Our old friend Matt Chambers becomes a new member in the Zhang Lab. Welcome, Matt!
[2015-06] Zhang Lab welcomes four new members, including Suhas Vasaikar, a Postdoctoral Research Fellow from Karolinska Institute; David Ma, a recent college graduate from Washington University in St. Louis; and two summer intern students, Kofi Amoah from Fisk University and Daniel Hong from Martin Luther King Magnet.
[2015-04] Dr. Zhang is a recipient of the 2015 CQS High Impact Award for the article, “Proteogenomic characterization of human colon and rectal cancer” [Zhang, et al. (2014) Nature, 513, 382-387].
[2015-03] IGP student Romell Gletten joined our group for a research rotation. Welcome, Romell!
[2015-01] Dr. Zhang is a recipient of the 2015 VICB Highly-Cited Article Award for the article, “Protein identification using customized protein sequence databases derived from RNA-Seq data” [Xiaojing Wang, et al. (2012) J. Proteome Res., 11, 1009-1017].
[2014-12] Jing’s paper Empowering biologists with multi-omics data: colorectal cancer as a paradigm has been published in Bioinformatics. In this article, we describe the application of the NetGestalt framework to provide a network-centric view of multidimensional cancer omics data, using colorectal cancer (CRC) as an example. The NetGestalt CRC portal is publicly available at the NetGestalt website.
[2014-11] We have a new opening for an Application Developer position. Please check the Positions page for more information.
[2014-10] Our collaborative study with Dr. Beauchamp’s group leading to the discovery of NFATc1 as an invasive driver in colon cancer has been published in Cancer Research in a paper entitled Nuclear factor of activated T-cell activity is associated with metastatic capacity in colon cancer.
[2014-10] Our review article Leveraging the complementary nature of RNA-Seq and shotgun proteomics data has been published in Proteomics. Congratulations, Xiaojing and Qi!
[2014-09] Dr. Zhang received the 2014 DBMI Outstanding Educator Award.
[2014-08] DBMI postdoctoral trainee Jason Castellanos, MD, joined our group for a research rotation this Fall. Welcome, Jason!
[2014-08] QCB student Michael Greer joined our group for a research rotation. Welcome, Michael!
[2014-08] Peter has created eight excellent tutorial videos for NetGestalt. They are available from the NetGestalt homepage. Try the “What is NetGestalt” video now!
[2014-07] Xiaojing received the best poster award from the 3rd Mass Spectrometry Special Interest Group (MS-SIG) meeting at ISMB 2014, Boston. Congratulations, Xiaojing!
[2014-07] The paper Proteogenomic characterization of human colon and rectal cancer is now online in Nature. Congratulations to all lab members who participated in this study and our collaborators!
[2014-07] Dr. Yongchao Dou joined the group as a Postdoctoral Research Fellow. Welcome to the group!
[2014-07] Mr. Peter Straub joined the group as a Bioinformatics Systems Engineer. Welcome to the group!
[2014-07] Dr. Xiaojing Wang has been promoted to Research Instructor of Biomedical Informatics effective July 1, 2014. Congratulations, Xiaojing!
[2014-05] Xiaojing’s review article Integrating genomic, transcriptomic and interactome data to improve peptide and protein identification in shotgun proteomics has been published in Journal of Proteome Research. Congratulations!
[2014-05] Congratulations to Jing (co-first author) and our wet-lab collaborators at the Vanderbilt Conte Center on the paper An open-source analytical platform for analysis of C. elegans swimming induced paralysis published in the Journal of Neuroscience Methods. The paper describes SwimR, an R-based suite that calculates, analyzes, and plots the frequency of C. elegans swimming behavior over time. It places a particular emphasis on identifying paralysis and quantifying the kinetic elements of paralysis during swimming. SwimR is available at Bioconductor.
[2014-03] IGP student Michael Bray will be doing a research rotation in our group. Welcome!
[2013-11] Jing’s paper Deciphering genomic alterations in colorectal cancer through transcriptional subtype-based network analysis has been published in PLoS One. This work demonstrated that integrating transcriptional subtype and signaling network analyses can help reveal subtype-specific signaling network alterations and provide insights into the development of personalized therapeutic strategies. Congratulations, Jing!
[2013-11] Congratulations to XiaoJing (co-first author) and our wet-lab collaborators at the Jim Ayers Institute for Precancer Detection and Diagnosis on the Cancer Research paper Proteogenomic analysis reveals unanticipated adaptations of colorectal tumor cells to deficiencies in DNA mismatch repair.
[2013-10] Jing received the 2013 DBMI Research Staff Excellence Award. Congratulations!
[2013-09] Xiaojing’s paper customProDB: an R package to generate customized protein databases from RNA-Seq data for proteomics search has been published in Bioinformatics. Congratulations, Xiaojing!
[2013-09] Qi’s paper RNA-seq data analysis at the gene and CDS levels provides a comprehensive view of transcriptome responses induced by 4-hydroxynonenal has been published inMolecular BioSystems. Congratulations, Qi!
[2013-08] Congratulations to Jing and our wet-lab collaborators in the Massion Lab on the Clinical Cancer Research paper “Integrative genomics analysis identifies candidate drivers at 3q26-29 amplicon in squamous cell carcinoma of the lung“. Using an integrative computational analysis followed by experimental validation, the study identifed three candidate driver genes (SENP2, DCUN1D1 and DVL3) that could potentially stratify patients with lung squamous cell carcinoma into subgroups with different response to adjuvant chemotherapy.
[2013-07] NetGestalt was highlighted in the VUMC news.
[2013-06] NetGestalt has been published in Nature Methods. NetGestalt is a novel data integration framework that allows simultaneous presentation of large-scale experimental and annotation data from many sources in the context of biological networks to facilitate data visualization, analysis, interpretation and hypothesis generation. Congratulations, Zhiao and Jing!
[2013-06] In an interview with the eProtein newsletter of NCI, Dr. Zhang expressed his view on proteogenomic data integration.
[2013-05] Two undergraduate students joined the group through the DBMI summer internship program. Liron Ganel is a rising senior majoring in Biochemistry, Computer Science, and Mathematics at the Indiana University, Bloomington. Derek Shyr is a returning summer intern from the Washington University in St. Louis. Welcome both Liron and Derek!
[2013-05] Congratulations to Jing on the new Nucleid Acids Research paper WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. This is an update from a previous publication on WebGestalt in the same journal in 2005. For the last 7 years, WebGestalt data holdings have grown substantially to satisfy the requirements of users from different research areas. The current version of WebGestalt supports 8 organisms and 201 gene identifiers from various databases and different technology platforms. Meanwhile, by integrating functional categories derived from centrally and publicly curated databases as well as computational analyses, WebGestalt has significantly increased the coverage of functional categories in various biological contexts including Gene Ontology, pathway, network module, gene–phenotype association, gene–disease association, gene–drug association and chromosomal location, leading to a total of 78,612 functional categories. Finally, new interactive features, such as pathway map, hierarchical network visualization and phenotype ontology visualization have been added to WebGestalt to help users better understand the enrichment results. WebGestalt can be freely accessed through http://www.webgestalt.org or http://bioinfo.vanderbilt.edu/webgestalt/ .
[2013-04] Dr. Zhang has received a three-year contract award from the SAIC Frederick, Inc. This contract award will support the continuous development of NetGestalt, a web application for integrating multidimensional omic data over biological networks. NetGestalt will be used to build a cancer omics data browser for data generated from the NCI TCGA and CPTAC projects.
[2013-04] Qi’s MCP paper was highlighted on the Discoveries Featured of the VICB communications.
[2013-04] Qi’s paper Integrative omics analysis reveals the importance and scope of translational repression in microRNA-mediated regulation has been published in Molecular & Cellular Proteomics. Through integrative analysis of miRNA, mRNA and protein expression data in nine colon cancer cell lines, we found that translational repression was involved in 58% and played a major role in 30% of all predicted miRNA-targeted interactions. Moreover, sequence features known to drive site efficacy in mRNA decay were generally not applicable to translational repression. Congratulations, Qi!
[2013-02] Dr. Qi Liu has been promoted to Research Assistant Professor of Biomedical Informatics effective February 1, 2013. Congratulations, Qi!
[2013-01] Jerome’s paper GLAD4U: deriving and prioritizing gene lists from PubMed literature has been published in BMC Genomics. Congratulations!
[2012-08] DBMI student Abigail Lind will be doing a research rotation in our group during the Fall semester. Welcome!
[2012-08] Mingguang’s paper a network-based gene expression signature informs prognosis and treatment for colorectal cancer patients has been published in PLoS One. Congratulations!
[2012-07] Derek Shyr, a rising sophomore at the Washington University in St. Louis joined the group for a summer internship. Welcome, Derek!
[2012-06] Amber Shindhelm, a rising senior majoring in Mathematics and Computer Science at Vanderbilt joined the group for a summer internship. Welcome, Amber!
[2011-11] Xiaojing’s paper Protein identification using customized protein sequence databases derived from RNA-Seq data has been published in Journal of Proteome Research. Congratulations!
[2011-10] GLAD4U was one of the Show Off Your Apps Winners in the NLM’s first software development challenge on the Innovative Uses of NLM Information.
[2011-09] Mr. Jing Wang joined the group as a Bioinformatics Systems Engineer. Jing has a Master’s Degree in Bioinformatics from the Harbin Medical University in China. Welcome to the group!
[2011-09] Mingguang’s paper Semi-supervised learning improves gene expression-based prediction of cancer recurrence has been published in Bioinformatics. Congratulations!
[2011-06] Our paper Relating protein adduction to gene expression changes: a systems approach is featured on the cover of the July issue of the Molecular Biosystems. It is also highlighted in Vanderbilt’s Reporter and evaluated by the Faculty of 1000 (F1000). As stated in the F1000 evaluation, “These researchers used powerful systems biology strategies to uncover mechanisms linking adduction of individual proteins by 4-hydroxynonenal to global changes in gene expression that accompany cellular exposure to this noxious lipid-derived electrophile. …… This paper highlights the capability of integrative approaches to generate new hypotheses to explain the pathogenesis of diseases that involve toxic electrophiles”. A pdf file of the paper with the Molecular Biosystems cover can be downloaded here.
[2011-06] Dr. Qi Liu joined the group as a visiting scholar. Dr. Liu is an Associate Professor of Bioinformatics at the Shanghai Jiaotong University in China. Welcome to the group!
[2011-06] Mindy is coming back to intern with us this summer, welcome back!
[2011-05] A research paper Relating protein adduction to gene expression changes: a systems approach has been published in Molecular Biosystems. The software Netwalker described in the paper can be downloaded from http://bioinfo.vanderbilt.edu/netwalker.
[2011-05] A software paper Fast network centrality analysis using GPUs has been published in BMC Bioinformatics. Source code for the GPU-based centrality computation algorithm described in the paper can be downloaded from http://bioinfo.vanderbilt.edu/gpu-fan.
[2011-05] Xiaojing received the Second Prize for her poster “Integrating RNA-Seq data improves protein identification in shotgun proteomics” presented at the 2011 VICC Systems Biology Retreat. Congratulations!
[2011-03] Jing’s paper A bioinformatics workflow for variant peptide detection in shotgun proteomics has been published in Molecular & Cellular Proteomics. Congratulations!
[2011-01] DBMI student Dong Wang will be rotating with our group for the Spring semester. Welcome!
[2010-11] Dr. Zhang received the High Impact Research Award from the Vanderbilt-Ingram Cancer Center for the work published in Molecular Systems Biology in 2009.
[2010-09] Dr. Jing Zhu joined the group as a Postdoctoral Research Fellow. Dr. Zhu just got her PhD in Bioinformatics from the University of Electronic Science and Technology of China. Welcome to the group!
[2010-06] Mindy Hong from the Emory University will be doing a summer intern in our group. Welcome!
[2010-05] Our paper Co-expression module analysis reveals biological processes, genomic gain, and regulatory mechanisms associated with breast cancer progression has been published inBMC Systems Biology. Source code for the module identification algorithm described in the paper can be downloaded from http://bioinfo.vanderbilt.edu/ice.
[2010-04] Jing has completed her postdoctoral training and has taken an Associate Professor position in the Shanghai Jiaotong University in China. Congratulations and all the best for the future endeavors!
[2010-04] CanProVar was highlighted in the NCI’s eProtein Newsletter and Vanderbilt’s Reporter.
[2010-04] Dr. Xiaojing Wang joined the group as a Postdoctoral Research Fellow. Dr. Wang got her PhD in Bioinformatics from the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences last summer. Welcome to the group!
[2010-03] Congratulations to Jerome on passing his Master’s thesis defense!
[2010-01] Dr. Mingguang Shi joined the group as a Postdoctoral Research Fellow. Dr. Shi got his PhD in Pattern Recognition and Artificial Intelligence from the University of Science and Technology of China last summer. Welcome to the group!
[2010-01] Jing’s paper CanProvar: a human cancer proteome variation database has been published in Human Mutation. Congratulations!
[2009-08] Jing’s paper Network-assisted protein identification and data interpretation in shotgun proteomics has been published in Molecular Systems Biology. Congratulations!
[2009-08] Naresh Prodduturi joined the group as a Bioinformatics Systems Engineer.
[2009-08] Bing Zhang was awarded an NIH grant from the National Institute of General Medical Sciences in support of the research project entitled “Systems Approach to the Biological Basis of Colon Cancer Metastases”.
[2009-06] Ruke Achoja from the Meharry Medical College will be doing a summer intern in our group.
[2009-05]Naresh Prodduturi, a new graduate from the bioinformatics MS program at the University of Texas at El Paso will be doing a summer intern in our group.
[2009-04] Dr. Zhang was awarded a pilot grant from the Silvio O. Conte Neuroscience Research Center, Vanderbilt University for a project entitled “Integrative Systems Biology Approaches to Elucidate Serotonin Network Modules and Their Regulators”.
[2008-08] DBMI student Cathy Derow will be rotating with our group for the Fall semester.
[2008-07] DBMI student Jerome Jourquin joined the group as a graduate student.
[2008-02] A research paper From pull-down data to protein interaction networks and complexes with biological relevance has been published in Bioinformatics.
[2008-01] DBMI students Jerome Jourquin and Peter Straub will be rotating with our group for the Spring semester.
[2007-11] The paper Proteomic parsimony through bipartite graph analysis improves accuracy and transparency was highlighted in the Reporter.
[2007-08] A research paper Proteomic parsimony through bipartite graph analysis improves accuracy and transparency has been published in Journal of Proteome Research.
[2007-04] Dr. Jing Li joined the group as a Postdoctoral Research Fellow.
[2007-04] Dr. Zhang was awarded a pilot grant from the Center in Molecular Toxicology, Vanderbilt University for a project entitled “Computational prediction of protein adduction mediated transcriptional regulation”.
[2006-10] A research paper Detecting differential and correlated protein expression in label-Free shotgun proteomics has been published in Journal of Proteome Research.
[2006-06]Dr. Zhang joined the Department of Biomedical Informatics, Vanderbilt University.